The CRISPR library screening approach encompasses a wide range of species and can be applied to diverse research areas, including the functional analysis of non-coding RNAs, investigation of disease resistance mechanisms, exploration of cellular pathways, and in-depth dissection of cellular functions. Researchers can design customized sub-libraries tailored to their specific research objectives, even based on the results of omics analyses, to perform more precise functional screenings. Additionally, researchers can flexibly choose different types of libraries for screening based on the characteristics of the molecules of interest. Consequently, the selection of CRISPR libraries demonstrates considerable flexibility and adaptability.
Given the significance and robustness of CRISPR library screening, initiating and effectively advancing screening efforts has become a focal point for researchers. Therefore, this article introduces several common CRISPR screening design strategies for reference and guidance.
CRISPR Screening Based on Cell Lines
Drug Pressure Screening
Drug pressure screening constitutes a widely utilized strategy, with a particularly critical operational workflow. Using a CRISPR knockout library as an example, the design of such a screening experiment is elucidated in detail (Figure 1). When conducting either drug resistance screening or sensitivity screening, it is imperative to establish both a control group and a drug-treated experimental group to ensure the accuracy and reliability of the experimental results. The primary distinction between the two lies in the drug dosage: high-dose drugs are employed in resistance screening, whereas low-dose drugs are used in sensitivity screening.
Upon completion of the experiment, high-throughput sequencing technology is employed to conduct an in-depth comparison of the types and abundances of single guide RNAs (sgRNAs) between the two groups of cells. Comparative analysis allows for the identification of genes that, when knocked out, either enhance cellular resistance to the drug or increase cellular sensitivity to the drug. This methodology provides a robust tool for comprehending the mechanisms of drug action and cellular response.
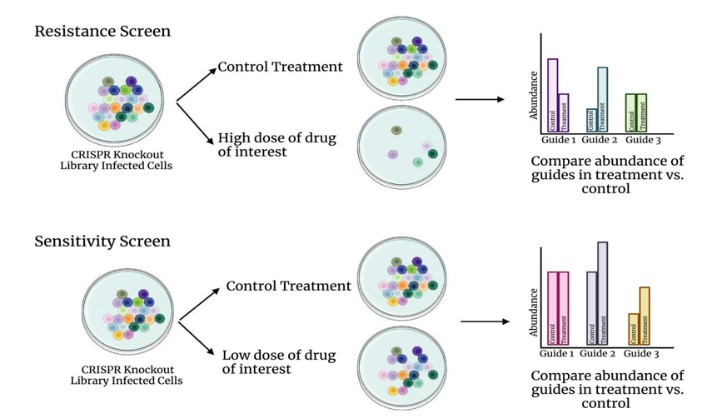
Figure 1. Schematic diagram of CRISPR drug screening experiment design
Take the Next Step: Explore Related Services
Cell Sorting
In addition to employing drug pressure screening methodologies, researchers can utilize fluorescence-activated cell sorting (FACS) for the selection of specific cell populations. This approach aims to identify functional sgRNAs by comparing the abundance of sgRNAs between different cell populations. The sorting process can be achieved based on the expression of fluorescent proteins by the cells (Figure 2) and can also employ fluorescent antibodies or specific fluorescent dyes to capture distinct cell groups (Figure 3). This scientific technique enables researchers to more precisely identify and analyze target cells, thereby providing robust support for subsequent investigations.
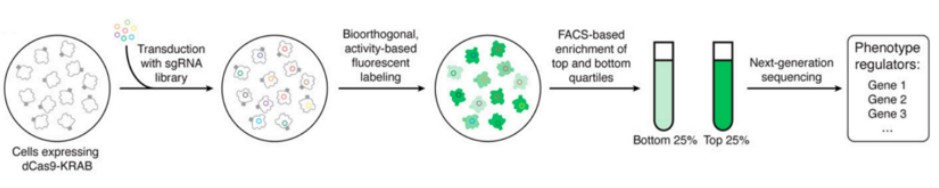
Figure 2 CRISPR selection based on fluorescent signal cell sorting
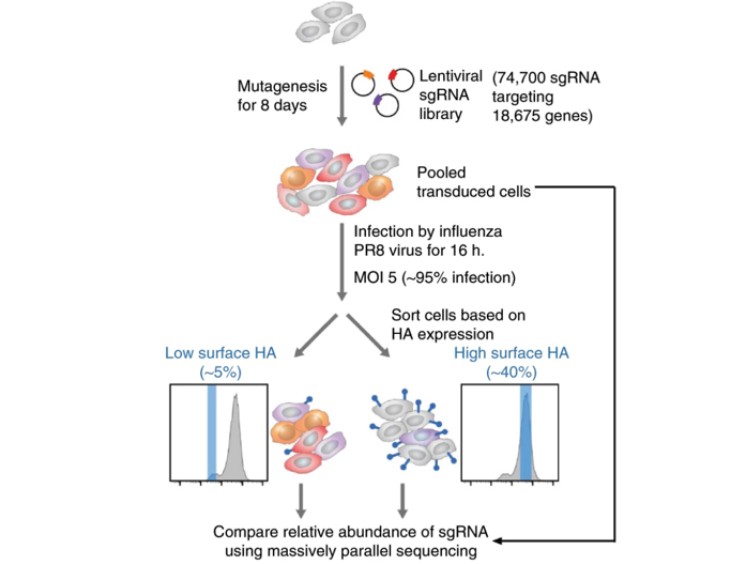
Figure 3 CRISPR screening based on fluorescent antibody capture of cells
Single-Cell CRISPR Screening
Single-cell CRISPR screening represents an innovative methodology that integrates CRISPR screening technology with single-cell transcriptome sequencing (scRNA-seq). This technique aims to elucidate gene function and genetic regulatory networks in a more comprehensive manner. Post-library screening, cells are labeled with specific barcodes to facilitate subsequent identification and analysis. Subsequent sequencing of both sgRNAs and messenger RNAs (mRNAs) allows for the comparison of sgRNA abundance and intracellular transcriptional changes at the single-cell level. This approach enables the precise correlation of “genotype-gene expression-phenotype” within individual cells, thereby providing a robust tool for uncovering gene function and regulatory mechanisms (Figure 4).
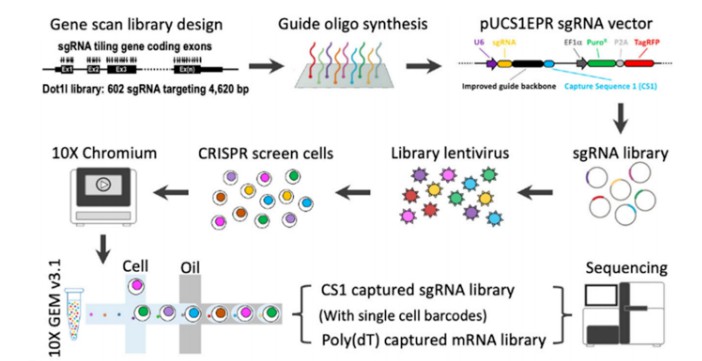
Figure 4 Schematic diagram of single-cell CRISPR selection process
Organoid-Based Screening
Organoids offer significant advantages over traditional two-dimensional culture systems in elucidating disease progression, homeostasis, and pathogenesis, thereby providing innovative approaches for disease diagnosis and treatment. In 2020, Henner F. Farin’s team in Germany successfully developed a tumor suppressor screening system based on CRISPR-Cas9 technology and human colonic organoids. This system is applicable to both in vitro culture and in vivo xenotransplantation platforms, offering personalized and precise therapeutic strategies for patients with highly heterogeneous colorectal cancer (Michels et al., 2020). The design of organoid screening processes closely resembles that of cell line-based screening (Figure 5), indicating that constructing an organoid screening platform does not substantially increase operational complexity.
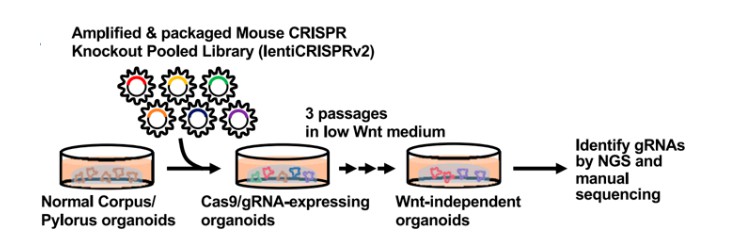
Figure 5 CRISPR screening of organoids
Screening Based on Animal Models
Animal models hold a pivotal role in contemporary life sciences research, particularly genetically engineered mice and rats. These models are indispensable for investigating gene functions, elucidating human physiological and pathological mechanisms, and facilitating novel drug development. The utilization of CRISPR screening technology in animal models enables researchers to precisely and efficiently identify critical functional genes (Figure 6). However, it is important to note that due to the limited number of available cells in animal screening processes, the size of the CRISPR libraries used is typically smaller to ensure adequate coverage. This limitation somewhat constrains the widespread adoption and application of this technology.
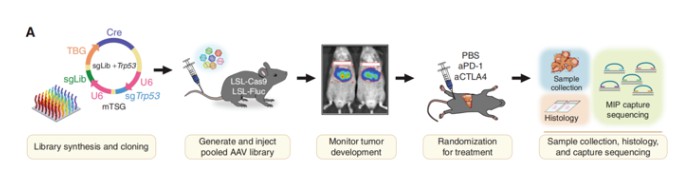
Figure 6 CRISPR in vivo screening based on animals
Screening Based on Cell and Animal Models
To address the limitations inherent in animal screening methods, researchers have developed an innovative combined approach integrating both cell-based and animal model screenings. Initially, standard cell line or organoid screening protocols are employed to identify candidate genes enriched in target cells or organoids. These enriched cells or organoids are then implanted into animal models to mimic conditions that closely resemble the physiological environment. By comparing the abundance of sgRNAs before and after implantation, it is possible to accurately identify key genes associated with specific phenotypes or functions (Figure 7). This integrative strategy, which combines cell and animal screenings, offers a more comprehensive and reliable methodology for investigating gene functions and disease mechanisms.
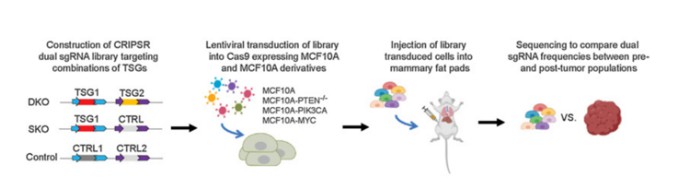 Figure 7 CRISPR screening in combination with cell and animal models
Figure 7 CRISPR screening in combination with cell and animal models
In recent years, the widespread application of CRISPR screening technology has increasingly demonstrated its significant value in the fields of life sciences and medicine. This technology has also given rise to numerous distinctive screening design strategies. Due to its high flexibility, CRISPR screening is poised to play a pivotal role in future research within related domains.
References
- Schleicher, E. M., & Moldovan, G. L. (2021). CRISPR screens guide the way for PARP and ATR inhibitor biomarker discovery. FEBS Journal, 3, 10.1111/febs.16217.
- Bumpus, T. W., Huang, S., Tei, R., & Baskin, J. M. (2021). Click chemistry-enabled CRISPR screening reveals GSK3 as a regulator of PLD signaling. Proceedings of the National Academy of Sciences of the United States of America, 118(48), e2025265118.
- Li, B., Clohisey, S. M., Chia, B. S., et al. (2020). Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nature Communications, 11(1), 164.
- Yang, L., Chan, A. K. N., Miyashita, K., et al. (2021). High-resolution characterization of gene function using single-cell CRISPR tiling screen. Nature Communications, 12(1), 4063.