Introduction to Acetylation in RNA
Histone acetylation has long been established as a mechanism for changing nucleosome structure. Direct links between transcription, histone acetylation, and the mechanisms by which histone acetylation is regulated have been formed in recent years. A substantial body of knowledge has accumulated regarding the mechanism(s) by which histone acetylation affects RNA polymerase II transcription. The impact of histone acetylation on the structure and function of rDNA, on the other hand, is poorly understood.

acRIP Sequencing: Introduction, Workflow, and Advantages
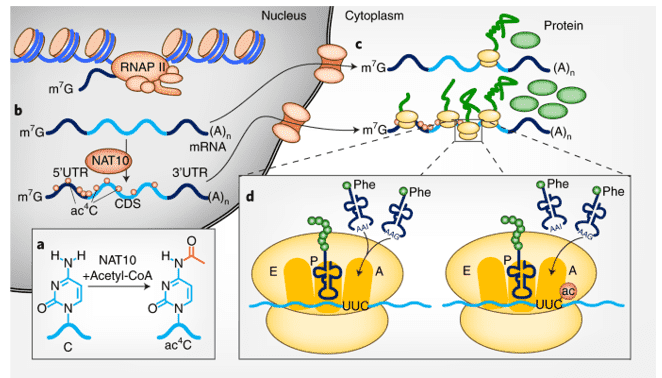
In eukaryotes and prokaryotes, ac4C is a conservative chemical modification. Previously, it was thought that ac4C only existed in tRNA and 18S rRNA; however, recent research has revealed that ac4C is also abundant in mRNA, with the majority of the enrichment sites appearing in coding sequences (CDS). The acetylation of N4-acetylcytosine by the ac4C modifying enzyme is known as ac4C acetylation. Ac4C acetylation is a new type of RNA modification that affects RNA stability and alternative splicing, as well as regulating gene expression, and is expected to become a novel epigenetic transcriptomics approach. acRIP sequencing can reveal the full picture of ac4C modification in a variety of RNA molecules quickly. The technique utilizes acetylated modified fragments enhanced by RNA co-immunoprecipitation as raw materials, extracts mRNA containing ac4C modifications, and integrates high-throughput sequencing technology with bioinformatics analysis, allowing systematic studies of ac4C modification in the transcriptome. acRIP sequencing can analyze the distribution and changes of ac4C in the transcriptome in great detail, allowing researchers to better understand the role of RNA acetylation in life and disease, and allowing for more precise disease diagnosis and treatment.
The stages in the acRIP sequencing workflow are as follows: (1) Poly(A) RNA purification, (2) RNA fragmentation (100-200 nucleotides), (3) RNA immunoprecipitation with anti-ac4C antibodies or isotypic IgG control, (4) acetylated RNA elution and purification, (5) rRNA depletion, (6) in vitro transcription of acetylated RNA, and (7) library preparation and high-throughput sequencing
The acRIP sequencing bioinformatics analysis pipeline includes (1) genome mapping and functional annotation of all mapped reads, (2) ac4C peak calling and visualization, (3) motif analysis, (4) differential acetylation analysis, (5) GO/KEGG enhancement analysis of differential gene/peak, (6) correlation analysis between differential ac4C and differential gene, and (7) correlation analysis between mRNA alternative splicing and differential ac4C, (8) correlation analysis between Ribo-seq and acRIP-seq.
The benefits of acRIP sequencing, on the other hand, include the generation of thousands of ac4C-enriched transcribed areas, high-resolution ac4C transcriptome mapping, and low cost.
References:
Choi SH, Meyer KD. Acetylation takes aim at mRNA. Nature structural & molecular biology. 2018 Dec;25(12).Figure 1. N4-acetylcytidine presents in cellular mRNAs and promotes translation. (Choi, 2018)
Sas-Chen A, Thomas JM, Matzov D, et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020 Jul;583(7817).
Arango D, Sturgill D, Oberdoerffer S. Immunoprecipitation and sequencing of acetylated RNA. Bio-protocol. 2019 Jun 20;9(12).
Based on a study, histone acetylation and/or acetylation of UBF or one of the other elements of rDNA transcription may be involved in affecting these dynamic changes instantly. The benefits of acRIP sequencing, on the other hand, include the generation of thousands of ac4C-enriched transcribed areas, high-resolution ac4C transcriptome mapping, and low cost. Temperature causes a significant increase in Ac4 C, and acetyltransferase-deficient archaeal strains have temperature-dependent growth defects. Cryo-electron microscopy was used to examine the temperature-dependent distribution of ac4 C and its potential thermoadaptive role in wild-type and acetyltransferase-deficient archaeal ribosomes.
Read More: ribosome profiling